|
|
ABI Systems Biology Group: Research |
|---|
Gene regulatory networks in cancer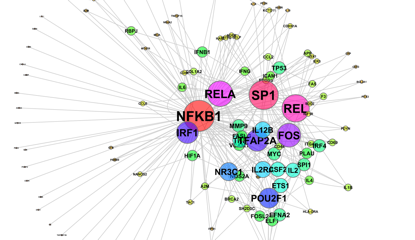 Developing
new cancer therapies or diagnostic tests relies on our comprehension of
the molecular switches regulating the progress of cancer. Although
advances have been made recently our knowledge about these molecular
switches remains incomplete. A new approach to understanding cancer
progression is to construct gene regulatory networks for specific cell
types. Gene regulatory networks are constructed from high-quality gene
array data from hundreds of disruptant and/or time-course experiments
in cultured cells. Gene regulatory networks are best described as
circuit diagrams showing cause and effect relationships between many
signalling molecules within cells. They ultimately identify master
regulators of gene expression represented by network ‘hubs’ from which
numerous signals emanate. Developing
new cancer therapies or diagnostic tests relies on our comprehension of
the molecular switches regulating the progress of cancer. Although
advances have been made recently our knowledge about these molecular
switches remains incomplete. A new approach to understanding cancer
progression is to construct gene regulatory networks for specific cell
types. Gene regulatory networks are constructed from high-quality gene
array data from hundreds of disruptant and/or time-course experiments
in cultured cells. Gene regulatory networks are best described as
circuit diagrams showing cause and effect relationships between many
signalling molecules within cells. They ultimately identify master
regulators of gene expression represented by network ‘hubs’ from which
numerous signals emanate.We are combining novel methods developed in our group with existing published methods for analysing gene expression data and applying them to data in breast, skin and colon cancer to uncover potential targets for investigation in the laboratory. Moreover, we are combining clinical information such as patient history, survival record, tumor grade and patient age, with molecular data to improve the predictive power of our approach. Key Publications:J.
Srividhya, M.A. Mourão, E.J. Crampin,
S. Schnell J.
Srividhya,
E.J. Crampin, P.E. McSharry, S. Schnell J. Wildenhain,
E.J. Crampin E.J. Crampin,
P.E. McSharry, S. Schnell E.J. Crampin, S.
Schnell, P.E. McSharry Collaborations:Cris Print group (Auckland), Mik Black (Otago), Santiago Schnell (University of Michigan) Recent Funding:
|
| Auckland Bioengineering Institute / Systems Biology Group / Research Projects |
|---|